(1)16S rDNA
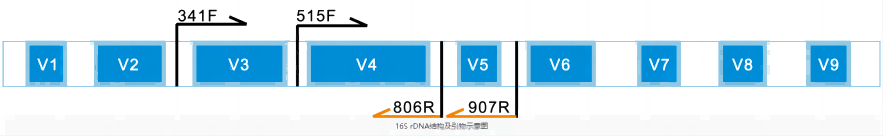
16S rDNA is the corresponding DNA sequence of the ribosomal 16S rRNA molecule in the prokaryotic genome, including conserved and hypervariable regions. By comparing the high-variable areas, different species can be distinguished and the degree of difference between them can be calculated. The 16S rRNA hypervariable region generally has 9 regions, each of which can reflect the differences between species to a certain extent, but the degree of reflection is different. Sequencing can be performed for one or two contiguous high-variable regions, and species screening can be performed against the database.
Sequencing based on the PacBio next-generation sequencing platform can also be used to study the differences in microbial diversity and community composition in combination with bioinformatics analysis. Compared with the second-generation sequencing, which only selects 1~2 high-variable regions for sequencing, the third-generation sequencing can obtain full-length 16S rDNA sequences, and more variant region information can improve the resolution of species identification, and can also improve the accuracy of community composition identification, and more realistically restore the microbial community structure in the sample.
(2)18S rDNA
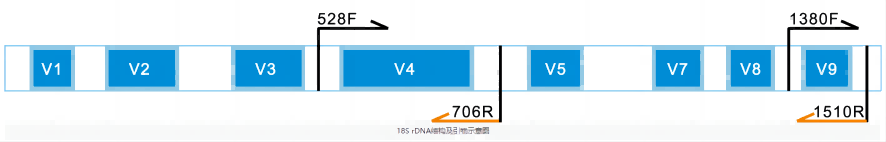
18S rDNA is the DNA sequence encoding a small subunit rRNA of ribosomes in eukaryotes, in which there are both conserved and hypervariable regions (V1-V9, no V6 region). Conserved regions reflect the genetic relationship between species, while variable regions can reflect the differences between species, which is suitable for the classification criteria of species and above. In the molecular ecology study of eukaryotes (such as plants, animals, fungi, protoplants, protozoa, etc.), the V4 region is used more, the database information is more complete, and the classification effect is better, which is a good choice for 18S rRNA gene analysis and annotation.
(3)ITS
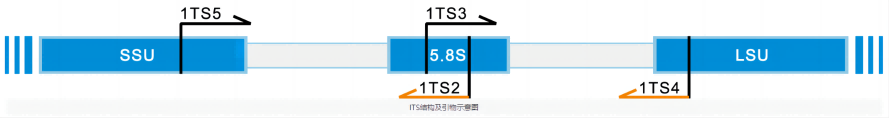
The full name of ITS is internal transcribed spacer, and on chromosomes, it is generally located between the RNA gene sequences of large and small subunits. In bacteria and archaea, ITS is located between the 16S and 23S rRNA gene sequences. In eukaryotes, there are two ITS fragments, ITS1 and ITS2. ITS1 is located between the 18S and 5.8S rRNA genes, while ITS2 is located between the 5.8S and 25S (plant) or 28S (animal) rRNA genes. ITS1 in eukaryotes corresponds to ITS in bacteria and archaea. ITS2 originated from the insertion of the 23S rRNA gene sequence of its ancestor.
In the study of the molecular ecology of fungi, the ITS region is currently the most commonly used sequencing target region, and we recommend using the ITS2 region for amplification, and ITS3 (5'-GCATCGATGAAGAACGCAGC-3') and ITS4 (5'-TCCTCCGCTTATTGATATGC-3') as amplification primers.
(4) Functional genes
The so-called functional microorganisms are a class of microorganisms that have received extensive attention in nature due to the importance of their functions, such as nitrifying bacteria, denitrifying bacteria, ammonia-oxidizing bacteria, sulfate-reducing bacteria, carbon-fixing bacteria, nitrogen-fixing bacteria, etc. Each functional microorganism may be taxonomically very different, but it has similar genes that enable it to perform the same function, so the genes that enable these functional bacteria to perform this specific function are called functional genes, such as nifH, nirS, nosZ, pmoA, dsrB, ureC, and phoD. For these functional genes, PCR universal primers were designed for high-throughput sequencing, which could analyze the functional community structure between samples, and combined with the environmental factors at each sampling point, the relationship between environmental factors and functional community structure could be analyzed at the same time.
The relationship between the genetic diversity of bacterial and fungal communities in the environment and their living environment has been gradually revealed in scientific research over the years, but there are still few studies on the genetic diversity of bacteriophages on the earth. There are a variety of highly conserved functional gene fragments in bacteriophages, so it is of great significance to design primers for conserved regions and identify them according to variable regions, design PCR universal primers, and perform high-throughput sequencing, which can explore the diversity of bacteriophages and clarify the structure and composition of bacterial communities in regulating ecosystems.