Metagenomic sequencing is based on the entire microbial community in a specific environment as the research object, which gets rid of the technical limitation that microorganisms are difficult to isolate and difficult to cultivate, and only needs to extract the total DNA of all microorganisms from the sample for sequencing. According to the different high-throughput sequencing platforms, it can be divided into metagenome (second generation) and metagenome (third generation).
Metagenomics (second generation): refers to the use of second-generation high-throughput sequencing technology to comprehensively sequence the DNA of microbial communities in samples, and to mine genetic diversity and molecular ecological information such as species composition, gene function, and metabolic pathways in samples through assembly, gene prediction, and gene function annotation.
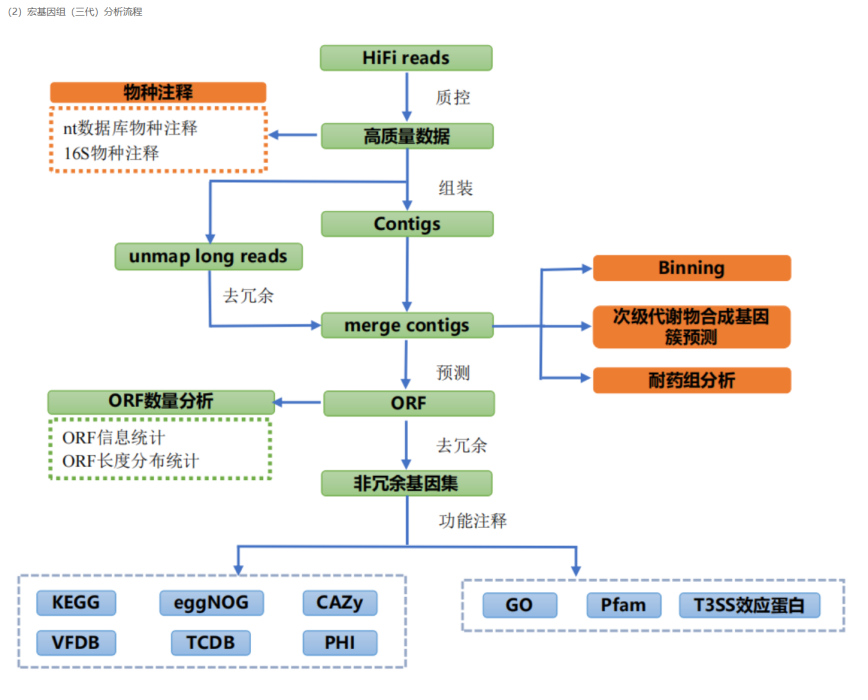
Metagenomics (third generation): refers to the use of third-generation high-throughput sequencing technology to comprehensively sequence the DNA of microbial communities in samples. Due to the development of next-generation sequencing technology, it can maintain long reads while taking into account accuracy, which can reduce some splicing errors, easily obtain high-quality sequence information, and improve the accuracy of genome assembly annotation and the resolution of microbial community identification.